Bioinformatics and Artificial Intelligence Platform
The Beijing Infectious Diseases Research Center has established a bioinformatics and artificial intelligence technology platform. The platform provides comprehensive support in terms of hardware, software, and personnel to meet the requirements for bioinformatics analysis and the development of artificial intelligence applications. Specifically, it includes:
1. Hardware Infrastructure
To meet the needs of bioinformatics and artificial intelligence technology development, the platform is equipped with a hardware support system consisting of high-performance CPU computing platforms, high-performance GPU computing platforms, and high-performance storage platforms. The platform can meet the requirements for genomic, transcriptomic (single-cell), proteomic, and metabolomic analysis, as well as the needs for building large language model knowledge bases, model fine-tuning, and agent development in clinical and research settings.
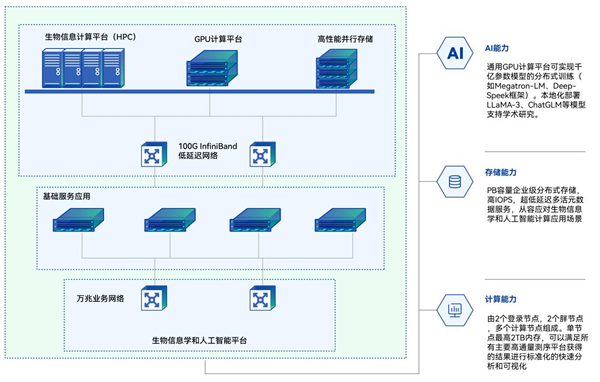
High-Performance Computing Platform for Bioinformatics (HPC)
GPU Computing Platform
High-Performance Parallel Storage
100G InfiniBand Low-Latency Network
Basic Service Applications
10 Gigabit Business Network
Bioinformatics and Artificial Intelligence Platform
AI Capability: Distributed training of billion-parameter models (e.g., Megatron-LM, Deep-Speek framework) can be realized using GPU computing platforms. Local deployment of models including LLAMA-3 and ChatGLM supports academic research.
Storage Capacity: Enterprise-level distributed storage with PB capacity, high IOPS, and ultra-low latency multi-active metadata services, efficiently handling life sciences and artificial intelligence computing application scenarios.
Computing Power: The system consists of 2 login nodes, 2 fat nodes, and multiple computing nodes, with each node equipped with up to 2TB of memory, capable of standardizing rapid analysis and visualization of results from all major high-throughput sequencing platforms.
(1) High-performance CPU computing platform (for bioinformatics analysis)
The computing nodes use AMD EPYC 7763 processors (64 cores, 2.45 GHz) with at least 1024 GB DDR4 memory per node, supporting collaborative computing between GPU and CPU. The platform provides a computational capability of no less than 25 TFLOPS, capable of parallel processing over 2000 threads of computing tasks.
(2) High-performance GPU computing platform (for artificial intelligence development)
The high-performance GPU computing platform, featuring 8 NVIDIA A100 Tensor Core GPUs (80GB VRAM) with NVLink interconnect technology, provides 312 TFLOPS of mixed precision computing power—suitable for deep learning model training (e.g., convolutional neural networks, Transformer), distributed training of billion-parameter models (e.g., Megatron-LM, DeepSpeek frameworks), and localized inference.
(3)High-performance Storage Platform
An enterprise-grade parallel storage system is used, with a total capacity of no less than 2PB, supporting a hybrid mode of object and block storage to meet the long-term archiving and rapid retrieval needs of multi-omics data. An enterprise-grade parallel storage system is used, with a total capacity of no less than 2PB, supporting a hybrid mode of object and block storage to meet the long-term archiving and rapid retrieval needs of multi-omics data.
2. Software Platform
(1) Bioinformatics Support Platform
This platform integrates open-source and self-developed analysis tools covering the full workflow of genomics, transcriptomics, proteomics, and metabolomics. It not only supports in-depth analysis of each individual omics but also enables cross-omics integration and joint exploration. In genomics, standard tools such as BWA, GATK, and self-developed TracePatho pathogen analysis and traceability system are deployed, supporting panoramic biomedical research from sequence variations to metabolic pathways. All tools are optimized for high-performance computing environments, ensuring efficient translation from raw data to biological insights.
(2) Local Deployment of Large Language Models for Artificial Intelligence
The local deployment of large language models for artificial intelligence has been achieved, supporting not only pre-trained general-purpose models but also deep optimization and custom development for the biomedical field. In terms of foundational models, open-source architectures such as LLaMA-3 and GPT-NeoX have been deployed. In terms of domain-specific enhancement, the platform integrates the PubMed literature database, clinical treatment guidelines, and omics databases for continuous training. Intelligent assistance has been implemented, from literature mining to experimental hypothesis generation, ensuring data privacy and security while providing cloud-level response speed, offering strong intelligent support for scientific innovation and clinical decision-making.
(3) National Clinical Infectious Disease Information Network System
By integrating third-party medical testing institutions (such as KingMed Diagnostics), the Ditan Hospital Infectious Disease Specialist Alliance, and sentinel hospitals, a nationwide clinical infectious disease information network system has been established. The system covers data from 31 provinces, 314 cities, and 4,518 medical institutions, encompassing 200 pathogens (including subtypes), with an annual sample volume exceeding 2 million cases. The data within this network is updated in real-time, with a data timeliness of T+2 days.
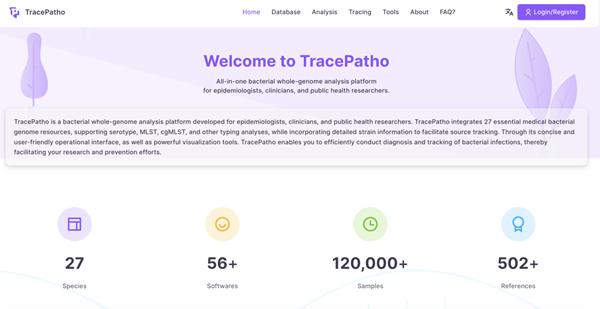
(4)Common Pathogenic Bacteria Analysis and Traceability System
The Common Pathogenic Bacteria Analysis and Traceability System (TracePatho) is a whole-genome analysis platform specifically designed for epidemiologists, clinicians, and public health researchers. It integrates 27 essential medical bacterial genomes and eight viral genome resources, supporting serotyping, MLST, cgMLST, and other typing analyses, while incorporating detailed strain information to facilitate source tracking. Through its concise, user-friendly interface and powerful visualization tools, TracePatho enables efficient diagnosis and tracking of microbial infections, thereby facilitating research and prevention efforts. (Website: https://tracepatho.com/)
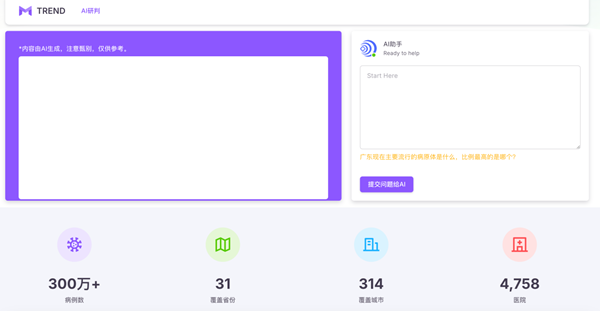
(5) Respiratory Infectious Disease Prevention and Control Expert Robot
Using DeepSeek as the foundational architecture for large models, the model is enhanced with the results of epidemiological research on 29 infectious diseases through Retrieval-Augmented Generation (RAG) technology, turning them into a knowledge base. The model is trained using data from over 3 million acute respiratory infectious disease cases, ultimately resulting in the development of a respiratory infectious disease prevention and control expert robot. This robot is capable of providing real-time Q&A services regarding infectious disease outbreaks and performing analysis and trend forecasting of epidemic developments.
3. Platform Development and Collaboration
The Bioinformatics and Artificial Intelligence (AI) Platform is a state-of-the-art research and development resource at the Beijing Infectious Diseases Research Center. We welcome partnerships with hospitals, universities, and research institutions for collaborative projects, data analysis, and shared innovation. For any inquiries, collaboration requests, or usage needs, please feel free to contact:
Email: nksunyamin@aliyun.com
Phone: +86 18622172239
PREV: Multi-Omics Platform

















































 京公网安备 11010502052111号
京公网安备 11010502052111号